import random
import pandas as pd
random_gamma = [random.gammavariate(1, 1) for x in range(1000)]
random_gamma = pd.Series(random_gamma)
random_gamma.plot(kind = "hist", bins = 50)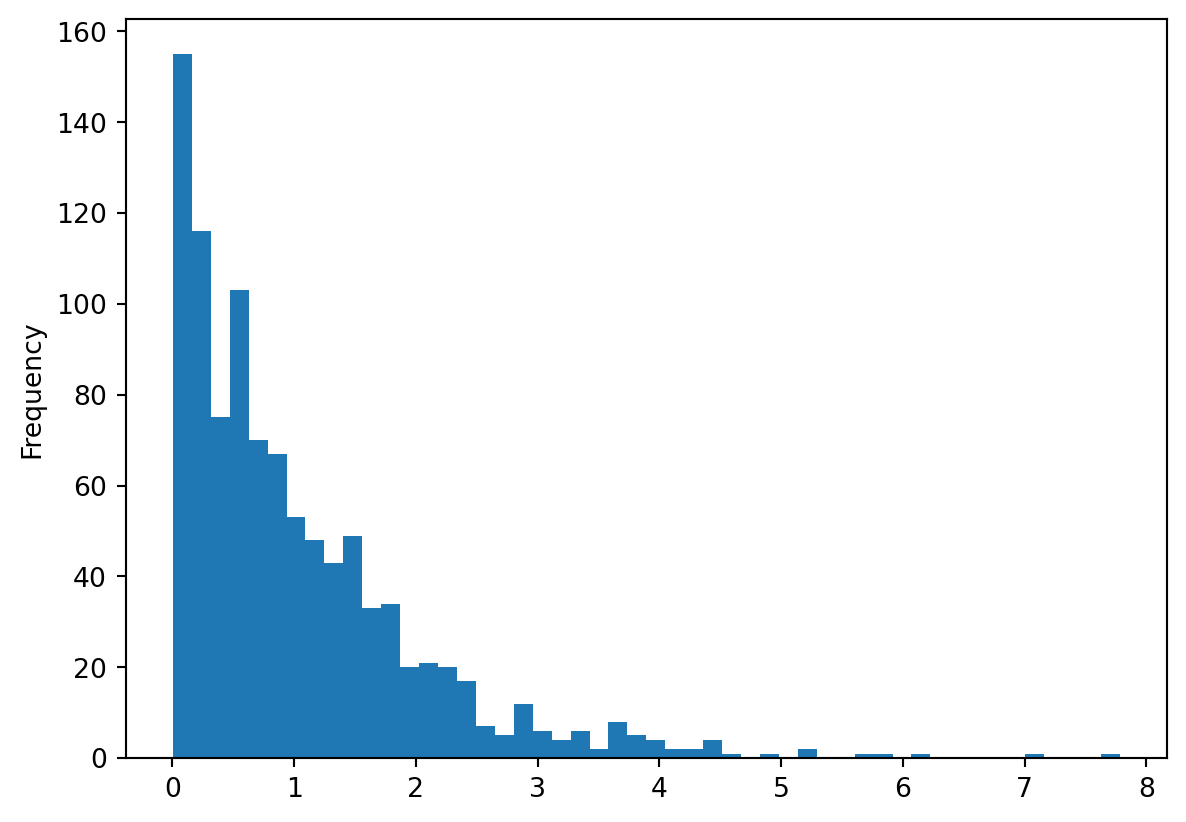
Es ist äusserst Zentral, Daten regelmässig und oft zu visualisieren. Die de facto standart Library hierfür in Python ist matplotlib. Diese Library kann man direkt ansteuern wie in diesem Tutorial beschrieben wird. Wir verwenden die library jedoch etwas anders: Dabei nutzen wir die Tatsache aus, dass sowohl pandas wie auch geopandas eingebaute Methoden (methods) haben um deren Inhalte zu visualisieren. Die Methode heisst in beiden Fällen .plot(), wie wir weiter unten noch sehen werden.
Listpandas vereinfacht das Visualisieren von Daten sogar soweit, dass es sich jewils lohnt seine Listen, Dictionaries usw. zuerst in eine Series oder DataFrame zu überführen um sie zu visualisieren (wie ich zum Beispiel in Zufallszahlen generieren jeweils gemacht habe).
import random
import pandas as pd
random_gamma = [random.gammavariate(1, 1) for x in range(1000)]
random_gamma = pd.Series(random_gamma)
random_gamma.plot(kind = "hist", bins = 50)
Weitere Beispiele zu Histogrammen aus Listen findet ihr im Kapitel Zufallszahlen generieren .
ListDas Visualisieren als Boxplot ist sehr ähnlich, man ersetzt "hist" lediglich durch "box. Eine komplette liste der möglichen Argumente für findet ihr hier: https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.Series.plot.html
random_gamma.plot(kind = "box")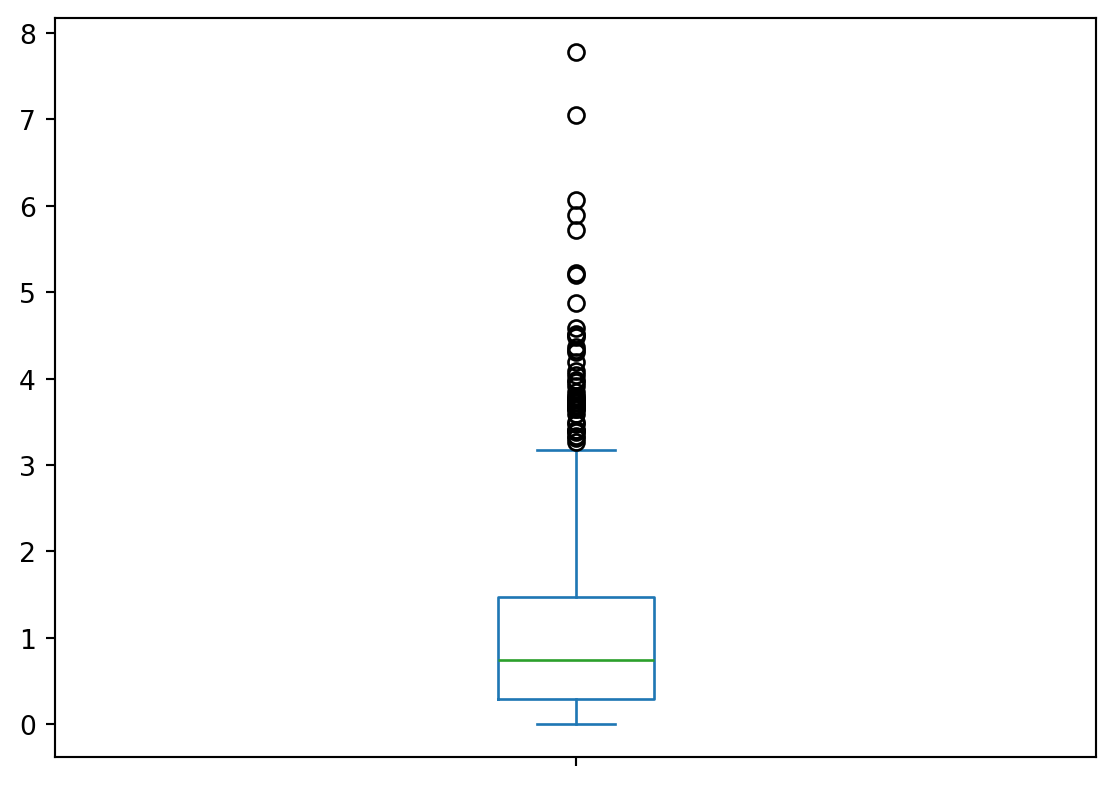
Um einen Scatterplot zu erstellen, braucht es eine DataFrame (eine Series reicht hierfür nicht aus).
zeckenstiche = pd.read_csv("data/zeckenstiche.csv")--------------------------------------------------------------------------- FileNotFoundError Traceback (most recent call last) Cell In[3], line 1 ----> 1 zeckenstiche = pd.read_csv("data/zeckenstiche.csv") File ~/miniconda3/envs/modul-agi/lib/python3.12/site-packages/pandas/io/parsers/readers.py:1026, in read_csv(filepath_or_buffer, sep, delimiter, header, names, index_col, usecols, dtype, engine, converters, true_values, false_values, skipinitialspace, skiprows, skipfooter, nrows, na_values, keep_default_na, na_filter, verbose, skip_blank_lines, parse_dates, infer_datetime_format, keep_date_col, date_parser, date_format, dayfirst, cache_dates, iterator, chunksize, compression, thousands, decimal, lineterminator, quotechar, quoting, doublequote, escapechar, comment, encoding, encoding_errors, dialect, on_bad_lines, delim_whitespace, low_memory, memory_map, float_precision, storage_options, dtype_backend) 1013 kwds_defaults = _refine_defaults_read( 1014 dialect, 1015 delimiter, (...) 1022 dtype_backend=dtype_backend, 1023 ) 1024 kwds.update(kwds_defaults) -> 1026 return _read(filepath_or_buffer, kwds) File ~/miniconda3/envs/modul-agi/lib/python3.12/site-packages/pandas/io/parsers/readers.py:620, in _read(filepath_or_buffer, kwds) 617 _validate_names(kwds.get("names", None)) 619 # Create the parser. --> 620 parser = TextFileReader(filepath_or_buffer, **kwds) 622 if chunksize or iterator: 623 return parser File ~/miniconda3/envs/modul-agi/lib/python3.12/site-packages/pandas/io/parsers/readers.py:1620, in TextFileReader.__init__(self, f, engine, **kwds) 1617 self.options["has_index_names"] = kwds["has_index_names"] 1619 self.handles: IOHandles | None = None -> 1620 self._engine = self._make_engine(f, self.engine) File ~/miniconda3/envs/modul-agi/lib/python3.12/site-packages/pandas/io/parsers/readers.py:1880, in TextFileReader._make_engine(self, f, engine) 1878 if "b" not in mode: 1879 mode += "b" -> 1880 self.handles = get_handle( 1881 f, 1882 mode, 1883 encoding=self.options.get("encoding", None), 1884 compression=self.options.get("compression", None), 1885 memory_map=self.options.get("memory_map", False), 1886 is_text=is_text, 1887 errors=self.options.get("encoding_errors", "strict"), 1888 storage_options=self.options.get("storage_options", None), 1889 ) 1890 assert self.handles is not None 1891 f = self.handles.handle File ~/miniconda3/envs/modul-agi/lib/python3.12/site-packages/pandas/io/common.py:873, in get_handle(path_or_buf, mode, encoding, compression, memory_map, is_text, errors, storage_options) 868 elif isinstance(handle, str): 869 # Check whether the filename is to be opened in binary mode. 870 # Binary mode does not support 'encoding' and 'newline'. 871 if ioargs.encoding and "b" not in ioargs.mode: 872 # Encoding --> 873 handle = open( 874 handle, 875 ioargs.mode, 876 encoding=ioargs.encoding, 877 errors=errors, 878 newline="", 879 ) 880 else: 881 # Binary mode 882 handle = open(handle, ioargs.mode) FileNotFoundError: [Errno 2] No such file or directory: 'data/zeckenstiche.csv'
zeckenstiche.plot("x", "y", kind = "scatter")--------------------------------------------------------------------------- NameError Traceback (most recent call last) Cell In[4], line 1 ----> 1 zeckenstiche.plot("x", "y", kind = "scatter") NameError: name 'zeckenstiche' is not defined
Um aus GeoDataFrames Karten zu machen verwendet man ebenfalls die .plot() Methode. Um wirklich schöne Karten mit Python herzustellen braucht man viel Übung. Für eine schnelle Visualisierung reicht aber die .plot() Methode.
import geopandas as gpd
zeckenstiche = gpd.read_file("data/zeckenstiche.gpkg")
wald = gpd.read_file("data/wald.gpkg")--------------------------------------------------------------------------- DataSourceError Traceback (most recent call last) Cell In[5], line 3 1 import geopandas as gpd ----> 3 zeckenstiche = gpd.read_file("data/zeckenstiche.gpkg") 4 wald = gpd.read_file("data/wald.gpkg") File ~/miniconda3/envs/modul-agi/lib/python3.12/site-packages/geopandas/io/file.py:294, in _read_file(filename, bbox, mask, columns, rows, engine, **kwargs) 291 from_bytes = True 293 if engine == "pyogrio": --> 294 return _read_file_pyogrio( 295 filename, bbox=bbox, mask=mask, columns=columns, rows=rows, **kwargs 296 ) 298 elif engine == "fiona": 299 if pd.api.types.is_file_like(filename): File ~/miniconda3/envs/modul-agi/lib/python3.12/site-packages/geopandas/io/file.py:547, in _read_file_pyogrio(path_or_bytes, bbox, mask, rows, **kwargs) 538 warnings.warn( 539 "The 'include_fields' and 'ignore_fields' keywords are deprecated, and " 540 "will be removed in a future release. You can use the 'columns' keyword " (...) 543 stacklevel=3, 544 ) 545 kwargs["columns"] = kwargs.pop("include_fields") --> 547 return pyogrio.read_dataframe(path_or_bytes, bbox=bbox, **kwargs) File ~/miniconda3/envs/modul-agi/lib/python3.12/site-packages/pyogrio/geopandas.py:261, in read_dataframe(path_or_buffer, layer, encoding, columns, read_geometry, force_2d, skip_features, max_features, where, bbox, mask, fids, sql, sql_dialect, fid_as_index, use_arrow, on_invalid, arrow_to_pandas_kwargs, **kwargs) 256 if not use_arrow: 257 # For arrow, datetimes are read as is. 258 # For numpy IO, datetimes are read as string values to preserve timezone info 259 # as numpy does not directly support timezones. 260 kwargs["datetime_as_string"] = True --> 261 result = read_func( 262 path_or_buffer, 263 layer=layer, 264 encoding=encoding, 265 columns=columns, 266 read_geometry=read_geometry, 267 force_2d=gdal_force_2d, 268 skip_features=skip_features, 269 max_features=max_features, 270 where=where, 271 bbox=bbox, 272 mask=mask, 273 fids=fids, 274 sql=sql, 275 sql_dialect=sql_dialect, 276 return_fids=fid_as_index, 277 **kwargs, 278 ) 280 if use_arrow: 281 meta, table = result File ~/miniconda3/envs/modul-agi/lib/python3.12/site-packages/pyogrio/raw.py:196, in read(path_or_buffer, layer, encoding, columns, read_geometry, force_2d, skip_features, max_features, where, bbox, mask, fids, sql, sql_dialect, return_fids, datetime_as_string, **kwargs) 56 """Read OGR data source into numpy arrays. 57 58 IMPORTANT: non-linear geometry types (e.g., MultiSurface) are converted (...) 191 192 """ 194 dataset_kwargs = _preprocess_options_key_value(kwargs) if kwargs else {} --> 196 return ogr_read( 197 get_vsi_path_or_buffer(path_or_buffer), 198 layer=layer, 199 encoding=encoding, 200 columns=columns, 201 read_geometry=read_geometry, 202 force_2d=force_2d, 203 skip_features=skip_features, 204 max_features=max_features or 0, 205 where=where, 206 bbox=bbox, 207 mask=_mask_to_wkb(mask), 208 fids=fids, 209 sql=sql, 210 sql_dialect=sql_dialect, 211 return_fids=return_fids, 212 dataset_kwargs=dataset_kwargs, 213 datetime_as_string=datetime_as_string, 214 ) File ~/miniconda3/envs/modul-agi/lib/python3.12/site-packages/pyogrio/_io.pyx:1239, in pyogrio._io.ogr_read() File ~/miniconda3/envs/modul-agi/lib/python3.12/site-packages/pyogrio/_io.pyx:219, in pyogrio._io.ogr_open() DataSourceError: data/zeckenstiche.gpkg: No such file or directory
Einfache Plots ohne anpassung:
wald.plot()--------------------------------------------------------------------------- NameError Traceback (most recent call last) Cell In[6], line 1 ----> 1 wald.plot() NameError: name 'wald' is not defined
Anpassung der Plot Grösse:
wald.plot(figsize = (5,5))--------------------------------------------------------------------------- NameError Traceback (most recent call last) Cell In[7], line 1 ----> 1 wald.plot(figsize = (5,5)) NameError: name 'wald' is not defined
Choroplethenkarte Karte
from matplotlib.colors import ListedColormap
my_cmap = ListedColormap(["green","lightgrey"])
wald.plot(column = "Wald_text", legend = True, cmap = my_cmap)--------------------------------------------------------------------------- NameError Traceback (most recent call last) Cell In[8], line 4 1 from matplotlib.colors import ListedColormap 3 my_cmap = ListedColormap(["green","lightgrey"]) ----> 4 wald.plot(column = "Wald_text", legend = True, cmap = my_cmap) NameError: name 'wald' is not defined
Mehrere Layers:
base = wald.plot(column = "Wald_text", legend = True, cmap = my_cmap)
zeckenstiche.plot(color = "Red", ax = base)--------------------------------------------------------------------------- NameError Traceback (most recent call last) Cell In[9], line 1 ----> 1 base = wald.plot(column = "Wald_text", legend = True, cmap = my_cmap) 2 zeckenstiche.plot(color = "Red", ax = base) NameError: name 'wald' is not defined
Weitere Informationen dazu findet ihr hier: https://geopandas.org/docs/user_guide/interactive_mapping.html
Vollautomatisch, ohne Anpassungen:
zeckenstiche.explore()Zeckenstiche Rot eingefärbt:
zeckenstiche.explore(color = "red")Zeckenstiche nach "accuracy" eingefärbt:
zeckenstiche.explore(column = "accuracy")Wald nach "Wald_text" eingefärbt (beachte, dass ich my_cmap weiter oben erstellt habe!):
wald.explore(column = "Wald_text", cmap = my_cmap)Zwei übereinander gelagerte Layers:
base = wald.explore(column = "Wald_text", cmap = my_cmap)
zeckenstiche.explore(m = base, color = "red")--------------------------------------------------------------------------- NameError Traceback (most recent call last) Cell In[10], line 1 ----> 1 base = wald.explore(column = "Wald_text", cmap = my_cmap) 2 zeckenstiche.explore(m = base, color = "red") NameError: name 'wald' is not defined
In folgenden Beispielen zeigen wir noch ein paar einfache Wege, wie ihr die Zeckenstichdaten visualisieren könnt.
import pandas as pd
import geopandas as gpd
import matplotlib.pyplot as plt
import descartes
wald = gpd.read_file("data/wald.gpkg")
zeckenstiche_full = pd.read_csv("data/zeckenstiche_full.csv")
geom = gpd.points_from_xy(zeckenstiche_full['x'], zeckenstiche_full['y'])
zeckenstiche_gpd = gpd.GeoDataFrame(
zeckenstiche_full,
geometry=geom,
crs = 2056)--------------------------------------------------------------------------- DataSourceError Traceback (most recent call last) Cell In[11], line 6 3 import matplotlib.pyplot as plt 4 import descartes ----> 6 wald = gpd.read_file("data/wald.gpkg") 7 zeckenstiche_full = pd.read_csv("data/zeckenstiche_full.csv") 9 geom = gpd.points_from_xy(zeckenstiche_full['x'], zeckenstiche_full['y']) File ~/miniconda3/envs/modul-agi/lib/python3.12/site-packages/geopandas/io/file.py:294, in _read_file(filename, bbox, mask, columns, rows, engine, **kwargs) 291 from_bytes = True 293 if engine == "pyogrio": --> 294 return _read_file_pyogrio( 295 filename, bbox=bbox, mask=mask, columns=columns, rows=rows, **kwargs 296 ) 298 elif engine == "fiona": 299 if pd.api.types.is_file_like(filename): File ~/miniconda3/envs/modul-agi/lib/python3.12/site-packages/geopandas/io/file.py:547, in _read_file_pyogrio(path_or_bytes, bbox, mask, rows, **kwargs) 538 warnings.warn( 539 "The 'include_fields' and 'ignore_fields' keywords are deprecated, and " 540 "will be removed in a future release. You can use the 'columns' keyword " (...) 543 stacklevel=3, 544 ) 545 kwargs["columns"] = kwargs.pop("include_fields") --> 547 return pyogrio.read_dataframe(path_or_bytes, bbox=bbox, **kwargs) File ~/miniconda3/envs/modul-agi/lib/python3.12/site-packages/pyogrio/geopandas.py:261, in read_dataframe(path_or_buffer, layer, encoding, columns, read_geometry, force_2d, skip_features, max_features, where, bbox, mask, fids, sql, sql_dialect, fid_as_index, use_arrow, on_invalid, arrow_to_pandas_kwargs, **kwargs) 256 if not use_arrow: 257 # For arrow, datetimes are read as is. 258 # For numpy IO, datetimes are read as string values to preserve timezone info 259 # as numpy does not directly support timezones. 260 kwargs["datetime_as_string"] = True --> 261 result = read_func( 262 path_or_buffer, 263 layer=layer, 264 encoding=encoding, 265 columns=columns, 266 read_geometry=read_geometry, 267 force_2d=gdal_force_2d, 268 skip_features=skip_features, 269 max_features=max_features, 270 where=where, 271 bbox=bbox, 272 mask=mask, 273 fids=fids, 274 sql=sql, 275 sql_dialect=sql_dialect, 276 return_fids=fid_as_index, 277 **kwargs, 278 ) 280 if use_arrow: 281 meta, table = result File ~/miniconda3/envs/modul-agi/lib/python3.12/site-packages/pyogrio/raw.py:196, in read(path_or_buffer, layer, encoding, columns, read_geometry, force_2d, skip_features, max_features, where, bbox, mask, fids, sql, sql_dialect, return_fids, datetime_as_string, **kwargs) 56 """Read OGR data source into numpy arrays. 57 58 IMPORTANT: non-linear geometry types (e.g., MultiSurface) are converted (...) 191 192 """ 194 dataset_kwargs = _preprocess_options_key_value(kwargs) if kwargs else {} --> 196 return ogr_read( 197 get_vsi_path_or_buffer(path_or_buffer), 198 layer=layer, 199 encoding=encoding, 200 columns=columns, 201 read_geometry=read_geometry, 202 force_2d=force_2d, 203 skip_features=skip_features, 204 max_features=max_features or 0, 205 where=where, 206 bbox=bbox, 207 mask=_mask_to_wkb(mask), 208 fids=fids, 209 sql=sql, 210 sql_dialect=sql_dialect, 211 return_fids=return_fids, 212 dataset_kwargs=dataset_kwargs, 213 datetime_as_string=datetime_as_string, 214 ) File ~/miniconda3/envs/modul-agi/lib/python3.12/site-packages/pyogrio/_io.pyx:1239, in pyogrio._io.ogr_read() File ~/miniconda3/envs/modul-agi/lib/python3.12/site-packages/pyogrio/_io.pyx:219, in pyogrio._io.ogr_open() DataSourceError: data/wald.gpkg: No such file or directory
Mit der Erweiterung seaborn lassen sich mit wenigen Zeilen Code eine Kernel Density rechnen und visualisieren. Siehe nachstehenden Code:
import seaborn as sns
f, ax = plt.subplots(1, figsize=(6, 6))
wald.boundary.plot(linewidth=0.1, ax=ax)
zeckenstiche_gpd.plot(color='red', linewidth=0.1, ax=ax)
sns.kdeplot(x = zeckenstiche_full.x, y= zeckenstiche_full.y,shade = False,n_levels = 10, cmap = "viridis", ax = ax)
ax.set_axis_off()
plt.show()--------------------------------------------------------------------------- ModuleNotFoundError Traceback (most recent call last) Cell In[12], line 1 ----> 1 import seaborn as sns 3 f, ax = plt.subplots(1, figsize=(6, 6)) 4 wald.boundary.plot(linewidth=0.1, ax=ax) ModuleNotFoundError: No module named 'seaborn'
Mit der Erweiterung contextily kann man sehr schnell und einfach Hintergrundkarten in den Plot einbinden. Dafür muss das GeoDataFrame vorher aber in WGS84 Koordinaten (EPSG 3857) konvertiert werden (mit to_crs, s.u.).
import contextily as ctx
f, ax = plt.subplots(1, figsize=(6, 6))
zeckenstiche_gpd.to_crs(epsg = 3857).plot(ax = ax)
ctx.add_basemap(ax)
ax.set_axis_off()
plt.show()--------------------------------------------------------------------------- ModuleNotFoundError Traceback (most recent call last) Cell In[13], line 1 ----> 1 import contextily as ctx 3 f, ax = plt.subplots(1, figsize=(6, 6)) 4 zeckenstiche_gpd.to_crs(epsg = 3857).plot(ax = ax) ModuleNotFoundError: No module named 'contextily'
Kernel Denisty und Hintergrundkarte können auch kombiniert werden:
lat = zeckenstiche_gpd.to_crs(epsg = 3857).geometry.x
lng = zeckenstiche_gpd.to_crs(epsg = 3857).geometry.y
f, ax = plt.subplots(1, figsize=(6, 6))
sns.kdeplot(x = lat,y = lng,shade = False,n_levels = 25, cmap = "viridis", ax = ax)
ctx.add_basemap(ax)
ax.set_axis_off()
plt.show()--------------------------------------------------------------------------- NameError Traceback (most recent call last) Cell In[14], line 1 ----> 1 lat = zeckenstiche_gpd.to_crs(epsg = 3857).geometry.x 2 lng = zeckenstiche_gpd.to_crs(epsg = 3857).geometry.y 4 f, ax = plt.subplots(1, figsize=(6, 6)) NameError: name 'zeckenstiche_gpd' is not defined